Selected publications:
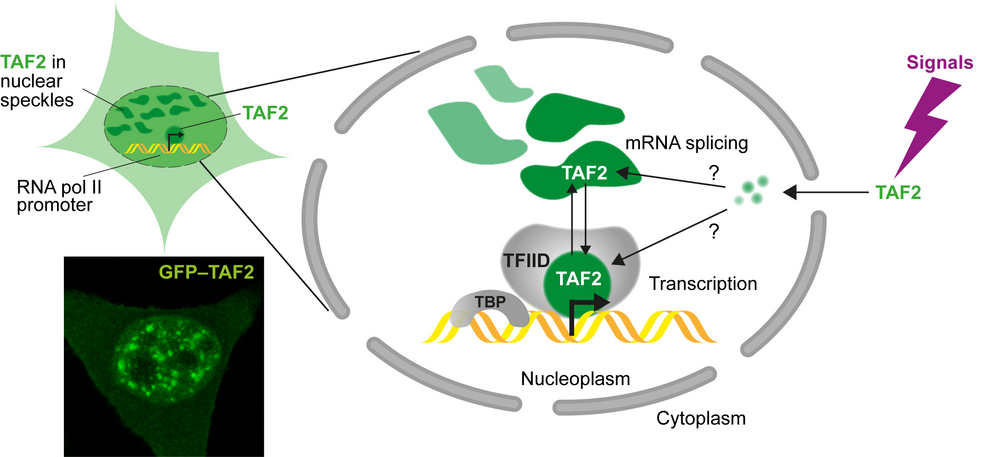
- Bhuiyan T.*,Arecco N., Mendoza Sanchez P.K., Kim J., Schwan C., Weyrauch S., Sheikh N., Prunotto A., Tekman M., Biniossek M.L., Knapp B., Koidl S., Drepper F., Huesgen P.F., Grosse R., Hugel T., Arnold S.J.* (2025), TAF2 condensation in nuclear speckles links basal transcription factor TFIID to RNA splicing factors. Cell Reports,Volume 44, Issue 5, 115616. DOI: https://doi.org/10.1016/j.celrep.2025.115616, *corresponding authors
Sheikh N., Koidl S., Bhuiyan T., Werner T.V., Biniossek M.L., Bonvin A., Lassmann S., Timmers H.T.M. (2021) Integrating quantitative proteomics with accurate genome profiling of transcription factors by greenCUT&RUN. Nucleic Acids Research,Volume 49, Issue 9, Page e49., DOI: https://doi.org/10.1093/nar/gkab038
Bhuiyan T. and Timmers H.T.M. (2019), Promoter recognition: putting TFIID on the spot., Trends in Cell Biology, Volume 29, Issue 9, pages 752-763., DOI: https://doi.org/10.1016/j.tcb.2019.06.004
Kapuria V., Rohrig U.F., Bhuiyan T., Borodkin V.S., van Aalten D.M., Zoet, V., Herr, W. (2016), Proteolysis of HCF-1 by Ser/Thr glycosylation-incompetent O-GlcNAc transferase:UDP-GlcNAc complexes. Genes & Development, Volume 30, pages 960-972., DOI: https://doi.org/10.1101/gad.275925.115
Bhuiyan T., Waridel P., Kapuria V., Zoete V., Herr W. (2015), Distinct OGT-binding sites promote HCF-1 cleavage. PLoS ONE, Volume10, Issue 8, e0136636, DOI: https://doi.org/10.1371/journal.pone.0136636
Lazarus M.B., Jiang J., Kapuria V., Bhuiyan T., Janetzko J., Zandberg W.F., Vocadlo D.J., Herr W., Walker S. (2013) HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science, Volume 342, No. 6163, pages 1235-1239, DOI: doi.org/10.1126/science.1243990