Efficient research data management (RDM) is imperative for pioneering research. At CIBSS — Centre for Integrative Biological Signalling Studies, we offer a comprehensive suite of tools that facilitate researchers in managing, sharing, and preserving their data in accordance with the highest FAIR principles (Findable, Accessible, Interoperable, Reusable). A selection of these tools have been adopted across a range of CIBSS research projects, while others, including OMERO and fredato, have been installed and customized by CIBSS members to address the specific needs of data management and collaboration within and beyond the research community. A comprehensive suite of resources is available for exploration, offering innovative solutions tailored to advanced biological signalling research.

Research Data Management Tools at CIBSS
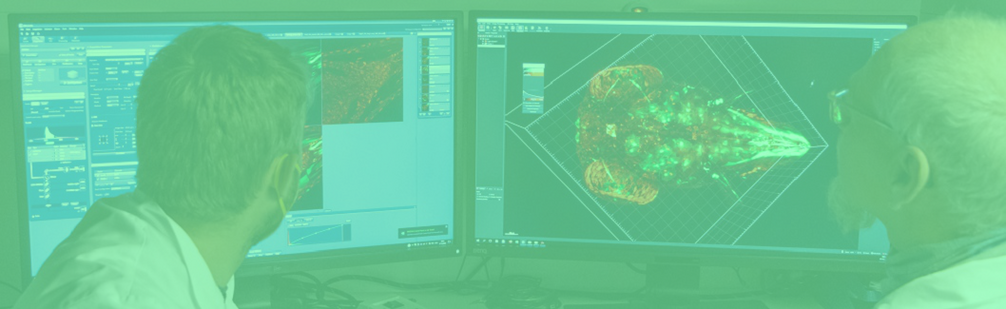
Omero — Open Microscopy Environment Remote Objects
CIBSS customized an microscopy imaging platform, called OMERO, to store, organize, and share raw microscopy data from publications, supporting over 150 file formats
Morefredato — Freiburg Research Data Tool
CIBSS customized fredato, a user-friendly tool for managing research data, synchronizing projects, and streamlining digital workflows with cloud storage and GitLab integration
MorebwSFS — Baden-Württemberg Storage For Science
bwSFS Storage at CIBSS provides secure, geo-redundant data storage for research groups, with customizable access for collaborative data sharing
MoreGalaxy
Galaxy is an open-source platform for FAIR data analysis, offering user-friendly workflows, interactive environments, and tools to manage, share, and reproduce data-driven research
MoreeLabFTW — Electronic Lab For The World
eLabFTW is a free, open-source electronic lab notebook designed to track experiments and manage labs securely and efficiently. At CIBSS, eLabFTW is used to manage single-molecule fluorescence data, including protein colocalization, cluster formation, and phase separation videos, along with analysis files.
MorePRIDE — PRoteomics IDEntifications Database
The PRIDE Archive is a public database for mass spectrometry proteomics data, including protein identifications, expression values, and supporting spectra. It is part of the ProteomeXchange consortium, enabling standardized data submission. At CIBSS, PRIDE stores and shares mass spectrometry data, such as dataset PXD047277, via the ProteomeXchange Consortium.
MoreGitHub
GitHub is a web-based platform for hosting and sharing free and open-source software. It provides version control, project discussion forums, DevOps tools for building and testing software, bug tracking, and documentation hosting, fostering collaboration. At CIBSS, GitHub is used to store and share protocols, scripts, and codes, supporting collaborative research and development.
Moredtool
dtool is a software suite for managing scientific data, providing a command-line interface and Python API to organize, transfer, and verify datasets across storage systems. At CIBSS, we use dtool with the dtool-lookup-gui and a lookup server for efficient, long-term management of research data, including single-molecule fluorescence tracking data, fluorescence videos of cells for protein colocalisation, cluster formation, and phase separation, along with all corresponding analysis files.
MoreResearch Infrastructure at the University of Freiburg include Core Facilities and Technology Platforms operated at the University and at the Medical Faculty, that are open for all researchers in Freiburg.
Research Infrastructure University of Freiburg
Research Infrastructure Medical Faculty











