Learning more about signals in cells needs new technologies: Researchers in the group of Prof. Barbara Di Ventura published a method, that can now select cells carrying two plasmids using a single antibiotic. Plasmids are small circular pieces of double-stranded DNA that can be taken up by cells in culture. This new technology called “SiMPl” enables researchers to control the function of several proteins independently. In contrast to previous technologies, this system selects modified cells using only one antibiotic, making the transfer of several plasmids more convenient and efficient. The study was released in the journal Nature Communications. The method is based on a cellular phenomenon called protein splicing. In signalling research, introducing genes encoding for proteins into cells in the lab helps to understand how they contribute to signaling pathways. Ultimately, researchers want to find out the role these genes and proteins play in organisms.


Simplifying cell selection with antibiotics
A new method developed by CIBSS researchers makes analyzing complex molecular pathways easier
This new procedure, developed for bacteria and mammalian cells, allows scientists to control the expression of two or more proteins independently, to study their interaction in a network of molecules. “It can easily be extended to be applied in other organisms, such as plants, provided one uses antibiotic selection,” Di Ventura, the senior author of the study and member of CIBSS – Centre for Integrative Biological Signalling Studies, comments. Additionally, using this method bacteria yield higher amounts of valuable chemicals that these introduced proteins helped create. Learn more about the procedure in the following graph and text.
Plasmids and Antibiotics
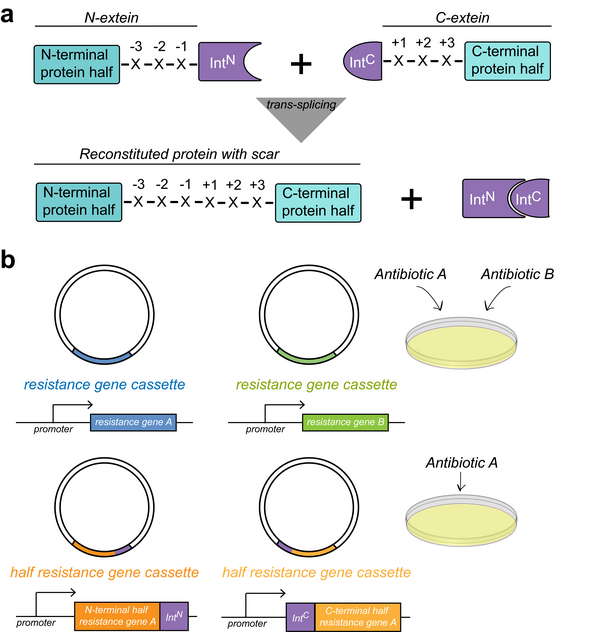
Genes coding for proteins are introduced in cells using plasmids. These circular DNA pieces contain the genes researchers want to introduce inside the cells, regulatory information for the cell and, additionally, an antibiotic resistance gene. By adding antibiotics such as ampicillin in the culture medium of these cells, only those that took up the plasmid carrying the resistance gene will be able to grow. Researchers can then be sure that all the cells that survived the antibiotic harbor the plasmid and can produce the protein encoded by the gene on the plasmid. Until now, to select cells containing two plasmids researchers had to use two different antibiotics. However, it is not always easy to find antibiotics that work well in the cell type of choice. Therefore, being able to use one single well-working antibiotic to introduce two plasmids is a great advantage.
Split inteins as bioengineering tools
The method developed by the group of Barbara Di Ventura uses so-called split inteins. Inteins are proteins that cut themselves out of a host protein in a process named protein splicing. During this process, chains of amino acids that constitute proteins are pasted together by a new bond between the two molecules that originally were flanking the intein. When inteins are split into two fragments they are called split inteins. To form a fully functional intein these fragments need to first bind to each other. Split inteins carry out a so-called trans-splicing reaction.For their ability to make fusions between two previously separate polypeptides, split inteins can be used to reconstitute a protein out of two dysfunctional parts. Therefore, they are extremely powerful bioengineering tools. In this work, Navaneethan Palanisamy, first author of the publication, split enzymes that confer resistance to the antibiotics in two parts. He then fused each part to one fragment of the split intein and inserted these fusion constructs into two separate plasmids. Once both plasmids are taken up by the cell and the proteins are synthetized, the split intein fragments find each other and perform the protein trans-splicing reaction, which results in the reconstituted, full-length, functional enzyme, which allow the cells to survive the antibiotic. The researchers named this method “SiMPl” because it is based on split intein-mediated enzyme reconstitution “and because it is indeed simple!” Di Ventura adds.
Implications for metabolic engineering and signalling studies
In collaboration with the group of Dr. Jung-Won Youn from the University of Stuttgart it was shown that, when using the SiMPl plasmids to express enzymes involved in a biosynthetic pathway, bacteria produce higher amounts of valuable chemicals. Furthermore, in collaboration with the group of Prof. Wolfgang Schamel from CIBSS, the researchers demonstrated that highly pure populations of mammalian cells double positive for two genes of interest could be obtained using the SiMPl plasmids conferring resistance to the antibiotic puromycin. Using this method, they could select functional Jurkat T cells expressing the a and b chains of the murine T cell receptor (TCR) using only puromycin.
Original publication:
Split intein-mediated selection of cells containing two plasmids using a single antibiotic.
Palanisamy N, Degen A, Morath A, Ballestin Ballestin J, Juraske C, Öztürk MA, Sprenger GA, Youn JW, Schamel WW, Di Ventura B.
Nat Commun. 2019 Oct 31;10(1):4967