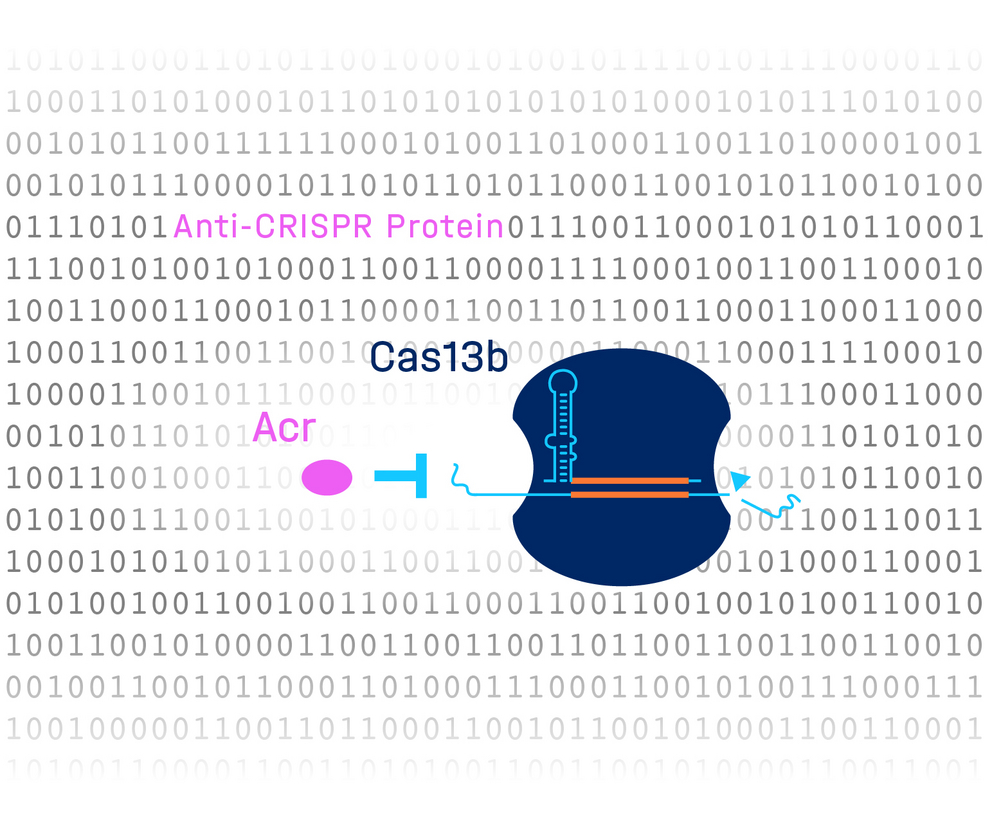
The nuclease Cas13b is associated with the CRISPR gene scissors. The enzyme, which degrades nucleic acids, has the potential to be used in future therapies of hereditary diseases to switch off unwanted genes. In addition, this nuclease is being researched as an antiviral agent in the fight against infections, as Cas13b can specifically intervene in the genetic material of viruses and render them harmless. An international research team aims to increase the safety and efficacy of future therapies. Led by bioinformatician Prof. Dr. Rolf Backofen from the Cluster of Excellence CIBSS - Centre for Integrative Biological Signalling Studies at the University of Freiburg and Prof. Dr. Chase Beisel at the Helmholtz Institute in Würzburg in cooperation with King Fahd University in Saudi Arabia, the researchers are looking for nuclease inhibitors that can regulate or prevent undesired side effects.


Deep learning helps improve gene therapies and antiviral drugs
Team led by CIBSS scientist Rolf Backofen develops algorithm that can improve gene therapies and antiviral drugs
Complex computational learning combined with a high-throughput screen revealed a novel anti-CRISPR protein that inhibits Cas13b. Image source: HIRI / Chase Beisel
In order to find natural nuclease inhibitors, CIBSS scientist Backofen from the Institute of Computer Science at the University of Freiburg and his team have now used Deep Learning for the first time to identify a protein that blocks the activity of Cas13b. Deep Learning is a subfield of machine learning and is suitable for examining large amounts of data for patterns and trends. The scientists present their results in the journal Molecular Cell.
In search of Acrs: Deep Learning applied for the first time
Scientists suspect that many undiscovered proteins exist that can block Cas13b, so-called anti-CRISPR proteins (Arcs). However, these are difficult to find. "Identifying them means finding a needle in a haystack, because Acrs don't resemble each other at all," explains Beisel, who is the lead scientist in the study in cooperation with Backofen and is head of the "Synthetic RNA Biology" department at the Helmholtz Institute for RNA-based Infection Research in Würzburg.
As a result, members of the research team have pushed the use of artificial intelligence to search for new acrs. "With the combination of our deep learning method 'DeepAcr' and the use of a high-throughput screen, we succeeded in discovering the new anti-CRISPR protein," Backofen says. "However, the millions of predictions made by our algorithm not only help to find anti-CRISPR proteins. The developed algorithm shows how neural networks open up effective solution possibilities even for complex problems."
Originalpublication
Wandera, K.G., Alkhnbashi, O.S., Bassett, H.V.I., Mitrofanov, A., Hauns, S., Migur, A., Backofen *, R., Beisel *, C.L. (2022): Anti-CRISPR prediction using deep learning reveals an inhibitor of Cas13b nucleases. In: Molecular Cell. DOI: https://doi.org/10.1016/j.molcel.2022.05.003 *:corresponding authors
Original press release



