The energy that a cell needs to function is mainly provided by its mitochondria: these structures convert energy from food through the so-called oxidative phosphorylation system (OXPHOS), an assembly of large molecules in the inner mitochondrial membrane. How the correct stoichiometry of the components of this vital machinery is achieved has remained elusive. Researchers at the Cluster of Excellence CIBSS at the University of Freiburg and at the ZMBH in Heidelberg have identified a novel regulatory mechanism that ensures the correct assembly of OXPHOS by receiving and processing signals from both, the nuclear and mitochondrial DNA.


Identification of a key molecular complex in mitochondria
Researchers from Freiburg and Heidelberg identify a crucial molecular mechanism that integrates information from nuclear and mitochondrial DNA
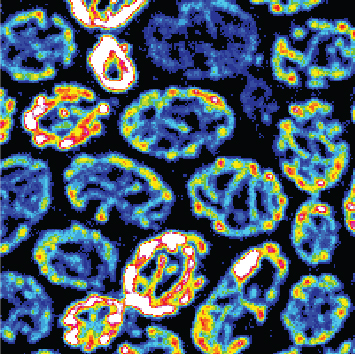
Fluorescence microscope image of the mitochondria of yeast cells. The membrane potential, which shows the activity of the mitochondria, is colour-coded. Image: Carlotta Peselj
The OXPHOS system consists of four molecular complexes that make up the electron transport chain and the enzyme ATP synthase. The correct stoichiometry between these components is essential, because otherwise oxygen radicals are formed, which are toxic to the cell. The current study, which was led by Prof. Dr. Nora Vögtle, Deputy Director of the ZMBH and member of the CIBSS, has now identified a sophisticated mechanism that synchronises OXPHOS assembly.
To study the regulation of OXPHOS, the researchers switched the food source of yeast from sugar to glycerol, forcing the cells to rapidly build up the OXPHOS system to avoid starvation. This metabolic rewiring induced the production of a previously uncharacterised protein, which the authors named Mra1. “We had previously identified Mra1 in proteomics approaches, but we did not know its function. The fact that it might play a role in metabolic rewiring spiked our interest and we wanted to uncover its molecular function,” says Vögtle.
Stop-and-go mechanism controls OXPHOS assembly
By combining several research techniques, including biochemical assays, proteomics, imaging and structural modelling, the authors identified a novel molecular assembly, which they termed MiRA and of which Mra1 is a part. “In relation to other cell components, MiRA really is gigantic,” describes Vögtle. MiRA controls the assembly of OXPHOS through a sophisticated stop-and-go mechanism: Mra1 acts as a molecular brake and stalls the assembly of one of the complexes of the electron transport chain. Two “Go” signals are then required for progression. “Intriguingly, these are encoded in the nuclear as well as in the mitochondrial DNA,” explains Vögtle. The two “Go” signals trigger the degradation of Mra1, which then allows the progression of OXPHOS assembly in a synchronised manner.
“The identification of MiRA was already very exciting, but the fact that we were then able to elucidate the function of this huge complex, its molecular mechanism and its physiological relevance, was really amazing,” says Vögtle. “But it also took us more than three years,” she adds. This mechanism could be one of the reasons why mitochondria have maintained their own genome, as it allows them to control OXPHOS assembly by integrating two different signalling sources. In addition, the assembly for other molecular complexes in the cell might be controlled by similar mechanisms, opening up new avenues of research.
Original publication
Daiana N. Moretti-Horten, Carlotta Peselj, Asli Aras Taskin, Lisa Myketin, Uwe Schulte, Oliver Einsle, Friedel Drepper, Marcin Luzarowski, F.-Nora Vögtle (2024). Synchronized assembly of the oxidative phosphorylation system controls mitochondrial respiration in yeast. In: Developmental Cell. DOI: 10.1016/j.devcel.2024.02.011
CIBSS-profile of Prof. Dr. Nora Vögtle



