During the first hours of an embryo's development, specialised molecules called pioneer transcription factors unravel parts of its DNA to activate the encoded genes. This has to happen at exactly the right times so that genes that are only needed at later stages of development are not activated too early. Biologists and physicists from the CIBSS Cluster of Excellence at the University of Freiburg now found what appears to be a fundamental mechanism for such a precise control of genes. Their findings, published in Nature Communications, contribute to our understanding of gene regulation in embryonic stem cells.


Kickstarting gene expression: it’s complicated
The same two transcription factors can enhance or inhibit each other, depending on the DNA sequence to which they bind, Freiburg researchers find
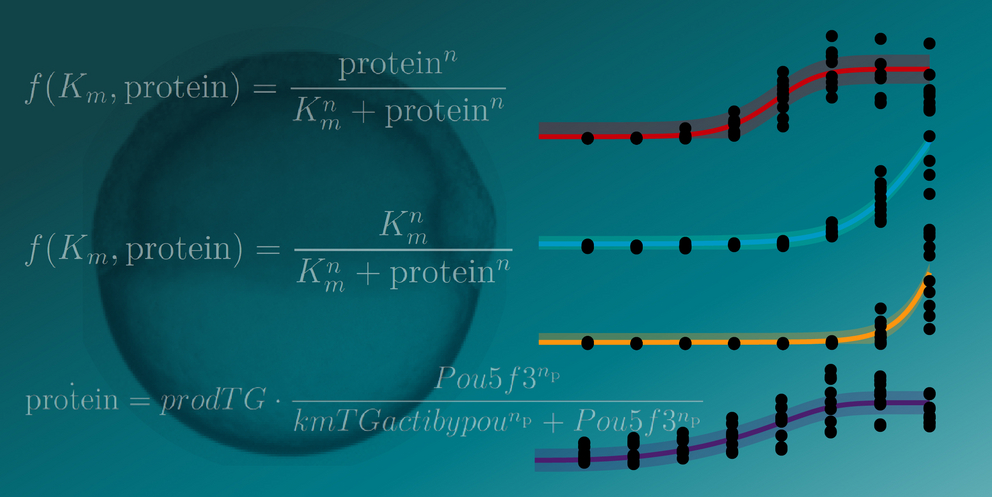
Collage showing a zebrafish embryo, graphs of measured gene expression levels (dots) and mathematical models (curves), and mathematical formulas from the publication. Collage: Michal Rössler / CIBSS
In a fertilised egg cell, some genes must be activated immediately, while others that trigger the differentiation of specialised cell types must still remain inactive. The current study, led by developmental biologist Dr. Daria Onichtchouk and theoretical physicist Prof. Dr. Jens Timmer, describes a previously unknown mechanism by which two activating transcription factors block each other actvities. The same factors that trigger the expression of early genes can thereby also prevent the premature expression of later genes.
The researchers show this for two zebrafish transcription factors, Pou5f3 and Nanog. The mammalian counterparts of these molecules, Oct4 and Nanog, are of high medical relevance because they are essential for the induction of induced pluripotent stem cells: a technology with high potential for future use in regenerative medicine, which was awarded the 2012 Nobel Prize.
“We knew that these transcription factors interact, but it was assumed that they could only make DNA more accessible, not less,” explains Onichtchouk. The researchers now show that there are indeed two possible modes of interaction: Pou5f3 and Nanog can either function synergistically to activate a gene, or antagonistically, inhibiting gene expression. “How they interact depends on the sequence of the DNA region they bind to,” says Onichtchouk. “This antagonistic interaction is what inhibits the expression of hundreds of genes during early stages of development.”
To reach this conclusion, the research groups of Onichtchouk and Timmer worked closely together, combining their expertise in developmental biology, genetics and mathematical modelling. Both scientists are members of the Cluster of Excellence CIBSS – Centre for Integrative Biological Signalling Studies at the University of Freiburg, which promotes such kinds of interdisciplinary projects.
Onichtchouk’s research group first compared the transcription factors’ effect on the expression of several hundreds of genes, using genetic assays. They then gave these data to the group of Timmer, asking them to model possible regulation mechanisms and without giving any further details on the specific factors they were investigating.
“The formulas we used were actually very simple and general, which makes the whole approach very elegant and me very happy,” says Jacques Hermes, who is doing his PhD in the group of Timmer and did a large part of the modelling for this study. He adds: “What is also very nice about this study is that we immediately had a way to validate the results yielded by our model using independent experimental data that we got.”
The model and experimental data show that Pou5f3 and Nanog synergistically activate those genes where they both act as activators, but have an antagonistic regulatory effect for those genes that can be activated by only one of them. In their publication, the researchers compare this interaction to two keys and a lock: If one of the key fits into the lock but cannot turn it, it will block the correct key.
“Now we know a lot more about the mechanisms by which transcription factors have context-dependent function,” Onichtchouk summarizes their finding. “This adds to the known complexity of gene regulation, but also brings us an important step closer to a complete understanding.”
Original publication
Aileen Julia Riesle, Meijiang Gao, Marcus Rosenblatt, Jacques Hermes, Helge Hass, Anna Gebhard, Marina Veil, Björn Grüning, Jens Timmer, Daria Onichtchouk. Activator-blocker model of transcriptional regulation by pioneer-like factors. In: Nature Communications (2023). DOI: 10.1038/s41467-023-41507-z
The original code from this study is freely accessible here: https://github.com/vandensich/zebrafish-minimodels
CIBSS-profile of Dr. Daria Onichtchouk
CIBSS-profile of Prof. Dr. Jens Timmer
The study was funded by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) and the Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, BMBF)



