Pioneering a new paradigm in drug discovery by leveraging biological understanding of how a virus interacts with its host, a team of researchers from USA, UK, France and Germany conducted a global protein phosphorylation analysis to determine mechanisms by which SARS-CoV-2 shifts cellular activity in infected cells. During the study, Prof. Robert Grosse and his group from the University of Freiburg’s Cluster of Excellence CIBSS discovered for the first time that virally infected cells exhibit filopodia, stringy arm-like extensions, which may be involved in speeding up viral spread throughout cells and tissues.
Phosphorylation, the addition of a phosphoryl group to a protein by an enzyme class called kinases, plays a pivotal role in the regulation of most cell processes including the production of the cytoskeleton, protein function, cell-to-cell communication, cell growth and cell death. The team led by Prof. Nevan Krogan, director of the Quantitative Biosciences Institute (QBI) at the School of Pharmacy at U.C. San Francisco (UCSF), senior investigator at Gladstone Institutes, determined that 40 of the 332 human proteins that interact with SARS-CoV-2 were significantly differentially phosphorylated. In addition, they identified 49 human kinases, out of a total of 518, that showed changes – either upregulation or downregulation – of phosphorylation activity.
The most strongly hijacked kinases include casein kinase II (CK2), kinases within the p38/MAP kinase (p38/MAPK) pathway, cyclin-dependent kinases (CDKs) and Phosphatidylinositol 5-kinase (PIKFYVE), all of which fall within a set of cell signaling pathways. Because kinases possess certain structural features, they are very druggable targets with more than 500 compounds commercially available or in development.
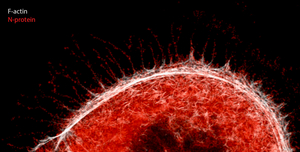
Interestingly, while the Freiburg research team from the Institute of Experimental and Clinical Pharmacology and the Institute of Virology was studying the impact of SARS-CoV-2 on CK2 in high-resolution imaging of virally-infected cells, they discovered actin-rich filopodia containing viral proteins. These images show that human CK2 and the viral N protein seem to co-localized within the filopodia, suggesting that SARS-CoV-2 hijacks CK2 and co-opts it into creating these tentacle-like protrusions that could possibly be manipulated to reach out to other cells or into the extracellular space and increase speed of infection. Further studies are needed to address this observation.
Other viruses including vaccinia, Ebola and Marburg take over the host cell cytoskeleton to promote egress and rapid cell-to-cell spread. However, in SARS-CoV-2 infected cells, the filopodia exhibit longer tentacles and branches, potentially enabling more aggressive transmission than other viral infection. “The distinct visualization of the extensive branching of the filopodia once again elucidates how understanding the biology of virus-host interaction can illuminate possible points of intervention in the disease,” Krogan comments in a press release by UCSF.
The quantitative study, ‘The Global Phosphorylation Landscape of SARS-CoV-2 Infection’ online now in Cell, builds upon a previously published novel “blueprint” of the 332 human proteins that interact with 27 SARS-CoV-2 viral proteins.
Researchers from Pedro Beltraos group at the EMBL’s European Bioinformatics Institute, Cambridge, UK, evaluated whether existing compounds would inhibit SARS-CoV-2 in infected cells in which specific kinases had been manipulated by the virus. The team triangulated changes in phosphorylation to specific protein kinase targets and identified 87 U.S. Food and Drug Administration (FDA) approved drugs, compounds in clinical trials or development. Based on an initial systematic review of these compounds, the researchers identified seven agents, primarily anti-cancer and inflammatory disease compounds that demonstrated antiviral activity in laboratory experiments: silmitasertib, gilteritnib, MAPK13-IN-1, SB203580, ralimetinib, apilimod and dinaciclib.
“Our data-driven approach for drug discovery has identified a new set of drugs that have great potential to fight COVID-19, either by themselves or in combination with other drugs, and we are excited to see if they will help end this pandemic.” Krogan further says.
Robert Grosse, who initiated and coordinated the Freiburg teams’ work, is professor at the CIBSS – Centre for Integrative Biological Signalling Studies of the University of Freiburg. He is director of the Institute of Experimental and Clinical Pharmacology and Toxicology. In his CIBSS project, he is researching signals of cells that are important for cytoskeleton structures such as the molecule actin – including kinases like CK2. This study was partly funded by CIBSS and the German Research Foundation (DFG).
Press release by UCSF: https://qbi.ucsf.edu/QCRGpressrelease
Press release by EMBL – EBI: https://www.ebi.ac.uk/about/news/press-releases/existing-drugs-can-prevent-sars-cov-2-hijacking-cells
Original publication: https://www.cell.com/cell/fulltext/S0092-8674(20)30811-4